Multiplexed Imaging: Using Multiplexed Tissue Imaging to Reveal the Spatial Biology of Cancer
On behalf of Cambridge Innovation Institute’s global web symposia series and our sponsor RareCyte, I’d like to welcome you to multiplex to tissue imaging the spatial biology of cancer. My name is Elizabeth Lamb and I’m the host and moderator for today’s event. Now. I would like to introduce our presenter for today, Dr. Sandro Santagata, MD, PhD, associate professor pathology, department of pathology, at Brigham and Women’s hospital. Welcome Dr. Santagata, the presenter ball is yours.
Dr. Sandro Santagata, MD PhD
Wonderful, hello everyone. Good morning, good afternoon, good evening to you. I’d like to start by thanking Elizabeth Lamb and my colleagues at RareCyte for organizing this webinar and inviting me to speak and of course thank you for attending. I look forward to your questions and your queries. I’m a neuropathologist at Brigham and Women’s Hospital, where I focus on brain tumor diagnosis. In addition, along with Peter Sorger, I co-lead the tissue imaging efforts at the laboratory of systems pharmacology at Harvard Medical School, where we work closely with groups, mostly in oncology but also in other disease areas where we use multiplex tissue imaging to study the spatial and molecular features of tissues from preclinical models all the way through on treatment, clinical trials.
Disclosures:
My disclosures are that I’ve been a co-investigator with RareCyte on a phase one small business technology transfer grant and STTR which recently transitioned to a phase two small business innovation research grant an SBIR and I’ll tell you about the Orion platform today that is the focus of that latter grant.
Objectives for multiplex tissue Webinar
Our objectives for this are, I’ll give you some background on multiplex tissue imaging the antibody-based version. We won’t speak today about spatial transcriptomic efforts. I’ll give you some examples, several examples of how we are using these methods, some insights, considerations and a little bit of a sense of the software infrastructure that we have in place. And of course, I’ll tell you about our experience working closely with RareCyte on the Orion platform, and I put a little asterisk here I’ll give you an overview of the larger arc of our work but this is obviously just touching the surface of some very exciting efforts that we are lucky to be participating in.
Team
I want to start by thanking our wonderful team at the LSP. Clearly, Peter, Jerry and Yu-an, they’re super integral to a lot of the efforts we have with RareCyte so a lot of the work that I’ll show you is theirs but lots of contributions from across the team and I’ll acknowledge those as we go through the presentation.
And I also want to point out that this work is really possible because of this kind of innovative environment we have at HMS, which is called the lab of systems pharmacology, LSP. Peter Sorger is the director. Laura Maliszewki’s the executive director and this is a multi-disciplinary space, where we bring together pathologists, oncologists, software engineers, data scientists, visualization experts, master administrators. It’s really a wonderful environment and I think this kind of provides the space where we can all meet, understand each other’s language and you know, make progress we hope towards, you know, innovations, towards human disease.
The Pathologist’s Toolbox
So as a pathologist, we have a toolbox and that’s been that our tools have gone, they’ve developed, some have been around for over a century and others are more recent in diagnosing a case. We start in the frozen section room where we get these artifacts written frozen sections with the ice crystal artifact that are H&E stained. We then get better sections later on and perform some immunochemistry and a lot of our work and diagnosis uses these modalities. Most recently obviously. they’re innovations in artificial intelligence applications and I won’t speak about those very powerful innovations today. Subsequent to that review, we started to layer in genomic information from FISH or copy number analyses as well as DNA sequencing and that pretty much covers our repertoire of tools. In addition, maybe some methylation profiling as well, most recently that allows us to tap into the tissues and provide diagnostic, prognostic, and predictive information. But we clearly know that there’s so much more information trapped in these tissues that we’re not unlocking and that would be really very beneficial to help us in drug development, in diagnosis, and prognosis so the field is trying to develop better techniques and one of those which I’ll speak about today is this multiple multiplex tissue imaging platforms.
Why Multiplexed Tissue Imaging
So why multiplex tissue imaging? Well you can see here in H&E and there’s a tremendous amount of information in those that as I mentioned AI approaches are starting to unlock but that the human eye has been has been doing quite a good job with for over a century and then we use immunistic chemistry but it’s a single marker IHC, sometimes two or three markers in some cases but mostly one marker, very helpful ancillary information but nonetheless it doesn’t provide us with the type of information that gives us deep mapping of tumors and tumor states. It’s certainly not very quantitative so it’s semi-quantitative at best. So multiplex tissue imaging methods have been developed to allow us to enumerate cell types and states. These basically cellular phenotypes and to map these positions, their positions and interactions in space. So when you have a platform that can give you 10, 15, 20, 30 markers or more on a single section, you can clearly identify which are normal epithelial cells, the tumor cells, the lymphocytes and the macrophages, what their states are, are they proliferating or secreting a certain ligand, etc etc, more and more information is available to us and then we know where they are relative to one another and it’s those types of maps that that our group and others are contributing to the human tumor atlas network which is a NCI funded effort to build a publicly available atlases with genomic data but also spatially resolve data into two and three dimensions.
T-CyCIF (Tissue Cyclic Immunofluorescence)
So, the method that we use at HMS, our principal method, although we’re increasingly using the Orion system, the method we use is one of a family of cyclic immunofluorescence methods and I’ll launch these two videos at the side here just to give you a sense of what those images look like. but then I’ll focus you over here where we basically take antibodies that are conjugated to fluorophores, we apply them to tissue sections. I’m not sure my video launched but maybe it’s going I’m sorry. So, we take these antibodies that are directly conjugated the fluorophores, we apply them to formalin and fixed paraffin-embedded tissue sections, we then provide a DNA stain, we do 4-channel imaging, sometimes 5-channel imaging using one of the RareCyte instruments, the HT3 which is a high capacity automated instrument, with high speed and high resolution. We then bleach the fluorophores and then apply new antibodies and then continue our cycles as many as are needed to generate our images. We generate large gigapixel images, they’re about anywhere from one centimeter square sometimes smaller but often larger, up to five centimeters squared and we do this by constructing and these composite images by stitching and registering many of the fields of view and then from these we extract single cell data and go off into computational space but also work to map back the data onto the slides. What I want to point out is that these are generally full slide images imaging and I want to make the case both with the Orion platform and with CyCIF, the value of doing whole slide imaging.
Generating Quantitative Data from Large Multiplexed Tissue Images
So we generate large amounts of data and we’ve had to develop a computational pipeline to handle this information. This is a presentation of a schematic of our MCMICRO pipeline which was recently published one week ago in Nature Methods and the tools are available freely in github. So, it’s a public open-source platform using either whole slides or TMAs. This is a modular pipeline that can then incorporate different algorithms at various stages. So, we can start with as I mentioned, whole slides are TMAs, it’s platform agnostic. We perform elimination correction and stitching as well as de-array of cores as needed. We can segment the cells, obviously that’s the main purpose of this using multiple different modules for segmentation, quantify the fluorescence level at single cell level and then generate these spatial feature tables which can be plugged into a whole range of different tools some of which are shown here. We’ve also generated a metadata standard, that will also be published in Nature methods shortly. So CyCIF data, Orion data can be analyzed by this, by this pipeline.
Sharing Insights with Digital Docents
I also want to point out another one of our tools and this is an online viewer for multiplex data that allows for narration of what one has seen in the images and a lot of multiplex data is abstracted into tiny’s and other plots which are very useful however it’s also very valuable to walk through the data and but it’s so complex that it needs a guide so we’ve been inspired by art galleries and how they have on online artwork narrated by digital docents and that’s what we’ve done here for multiplex imaging. This was recently published in Nature biomedical engineering about three weeks ago and if you want to see Orion data displayed in this system which we call Minerva, you can go to this link at the bottom of the images.rarecyte.com to explore really pretty lung cancers along normal lung specimens.
Multiplexed Imaging to Identify Markers of Combination Therapy Response
So, what are these platforms used for and I’ll get this is obviously the most traditional approach this is work from our good colleague Annina Farkkila at the University of Helsinki, who did much of this work when she was visiting us at the LSP. So in this project she was interested in trying to identify biomarkers of response to combined Niraparib park inhibitor and pembrolizumab anti PD-1 in the TOPACIO ovarian cancer trial. So simply what Annina and colleagues did was identify patients that responded and did not respond in this cohort and analyze their samples both genomically to find a mutational signature that ended up correlating with patient response as well as a spatial signature and this is an interaction between Phosphor-stat positive exhausted CD8 cells that express PD-1 and macrophage nearby macrophages that express PD-L1 or also tumor cells that express PD-L1. So this these inhibitory interactions were predictive of response to therapy so if a patient had either the signature or the response or both, there was a response and if the patient lacked both they did not respond to therapy and Jenn Guerriero, who also works very closely with us at the LSP, she’s an investigator at Brigham and Women’s, she’s doing a similar analysis now of the TOPACIO ovarian cancer trial. So the idea here is though, that in time, we’re going to develop these very important biomarkers and that they’re going to need validation and prospective biomarker trials and this is where we need to go at scale. This is data that was acquired by CyCIF but when we get to this, the scale that’s needed for these trials, 50, 100, 200 plus samples and prospectively acquired, that’s going to require a different approach that’s compatible with clinical workflows.
Relevant Spatial Features
Another example and this is maybe the opposite, but it’s also a spatial feature, it has to do with pure energy signaling in gliomas and what I mean by the opposite is that rather than be an inhibitory interaction that is possibly important here, this is a functionally cooperative interaction. So pure energy signaling, is this a really impressive extracellular signaling mechanism, where two enzymes CD39 and CD73 which are ecto enzymes, they take ATP from the extracellular space, metabolize it down to ANP, and then CD73 takes it further to adenosine, which is inhibitory to the immune system. What we find in glioblastoma looking across hundreds of samples is that there’s a functional interaction between these in a manner that resembles the vision of labor and by that I mean that the tumor cells express one of the enzymes CD73 and the immune cells mostly myeloid cells, express CD39 and they’re in proximity to one another so the 39 on the Myelo cells does the first part of the enzymatic reaction and the 73 on the tumor cells completes the reaction. So we find that this is not a random distribution of cells in the glioma microenvironment but this actually strong spatial correlation is shown by this plot here between the tumor cells that express 73 and the myeloid 39 expressing cells and that interaction correlates with worse response. So this is an example of a cooperative interaction in our field, as many of you on this webinar know, this is kind of our mandate now to try to find these functional ecosystems, these functional motifs present within tumors.
Multiplexed Imaging + Mouse GEMMs for studying the effects of combination therapies
A lot of our work is in human samples but one of our mandates for the STTR and the SBIR is to develop mouse panels that would allow us to probe model organisms, these genetically engineered mouse models. So in one of those projects with Tyler Jackson’s group, led by postdoctoral fellow Megan Berger and George Gaglia, who’s in my group. We’ve begun to image a range of mouse samples, these are lung adenocarcinomas that are initiated by an enzyme that’s expressed by a virus that then creates these tumor nodules in this genetically engineered mouse, multiple nodules across the whole lung so we image the entire specimen. We see a lot of heterogeneity ones that are hot and other ones nearby that are that are not hot, we can start to understand how neo antigens as well as vaccination and checkpoint therapies, influence the immune micro environment, the specific changes that they make as well as genetic manipulations within the tumors or within the micro environment and doing this we’re trying to build maps of the physical locations of the infiltrates, their interactions and their functional states. So that we can now move between mouse and humans. So this is a very important area of research, and this again is data from CyCIF but we have a lot of experience now with these antibodies which we’ll make available publicly shortly.
Multivariate Measurements Rather than Single Markers
I also want to point out that looking at the immune system is not the only use for these really innovative and powerful therapies, obviously we can look at the tumor intrinsic properties and what I’d like to point out is, you know something very basic, we can study cell proliferation in entirely new ways. The vast amount of the literature in research as well as clinical literature uses one marker for proliferative cells that’s Ki-67, but we, those of us that use it, know that this is rather fraught. It’s unclear how you quantify this, whether low expression or only high expression is positive. Whether you’re only counting tumor cells, we clearly know that there are small cells that might be immune cells or macrophages, lymphocytes, or macrophages, or fibroblasts that are caught up in endothelial cells, that get caught up in the counts so it’s not so easy to score. What Giorgio and Sheheryar Kabraji developed is a multivariate proliferation index. So using multiple markers, we start to probe whether cells are proliferating or not.
First of all, we can isolate away the immune cells and the stromal cells and just focus on the E-cadherin or cytokeratin positive tumor cells. In this example of breast cancer, we can see that Ki-67 is a marker that doesn’t mark all the proliferative cells but only ones that are later on in the cell cycle, actually in g2. We can see using other markers that we find cells that are Ki67 negative but positive for these proliferation markers. We can more readily identify cells that are not proliferating, study quiescent states in greater detail and we can find cells that are arrested. Mapping this information back onto this breast cancer specimen and looking at long range patterns, we see that there’s a lot of very clear autocorrelation between these proliferation states plus one being proliferative minus one arrested and zero non-proliferative and that they self-correlate in space likely due to all kinds of gradients which we’re starting to explore. We can also look at more local, at the 10 micron level interactions between cells and see that proliferative cells are more likely to be next to certain immune cells. In ER positive breast cancer and patterns that we see mostly recapitulated in other forms of breast cancer and even in ovarian cancer. And being even a little more innovative, is to look at extracting cell cycle dynamics from these snapshots of fixed human tissues. So what we do here is we isolate the proliferative cells, we then look at biomarker expression in those cells, eight cell cycle markers expanding various phases of the cell cycle, g1, sg2, and then we look at the correlation of those cell cycle markers in high dimensional space and then reduce them down into two dimensions and what they do is interestingly, they form a taurus, kind of reminiscent of a cycle, a cyclic process not surprising for the cell cycle. We can map the marker expression onto this reduced dimensionality representation and show that g1 markers are expressed in one portion of this whereas g2 markers are expressed in the next portion.
We can also develop a pseudo-time representation, where we start at one at ostensibly the g1 start point and follow the expression of markers over time with markers, endogenous markers, like cdt-1 dropping at g2 and going up at g2 etc and see how therapies influence these patterns both in terms of piling up in g1 or g2 a block or other changes which we see and report in this article that’s in Bioarchive and hopefully with these types of approaches, we can start to dose therapies to the desired effect to rather than to toxicity. And again this is where rapid methods are going to be very helpful both for clinical trials and hopefully for clinical management.
Multiplexed 3D Atlas of Colon Cancer
And what I’d like to do in the next several slides is give you a view of one colorectal cancer specimen with a deep dive into this one and this is just one of many that we’ve studied but I like showing one because it lets us focus in on some important features and you can find our presentation of this manuscript on Bioarchive. So what we did here is we took a sample from the cooperative human tissue network at Vanderbilt – it’s a colorectal cancer specimen, rather large about two centimeters by two centimeters, did serial sectioning through 106 plus specimens, we performed cyclic immunofluorescence on one section and then the neighboring section we stained with H&E and basically to allow us to build first generation three-dimensional atlases and to see how properties of the tumors change over space as well. We use the H&E and this is an important point that I also want to highlight for Orion, we use the H&E, which is an integral part of our analyses as is whole slide imaging as I mentioned earlier and hopefully this next few slides will highlight these points. Even though this is a moderately to highly poorly moderately to high moderate grade to high grade colorectal cancer specimen, it’s very heterogeneous. You can look at this and see that you know we have normal tissue here in ROI 1 we have moderately differentiated adenocarcinoma at different levels of depth of invasion here in ROIs 2, 3, and 4. We have very poorly differentiated, sheet-like growth of tumor cells in ROI 5, and then we have mucinous cells that are floating in mucin pools and ROI 6. And moreover even the invasive margins as I’ll show you in a few slides, are very different from another invasive margin A, B, and C. So one specimen is one label in principle in the pathology diagnosis but very heterogeneous internally so requiring review of the H&E and integration of multiplex data into that framework that we have.
Disease Relevant Morphologies in H&E Are Encoded as Hyperdimensional Patterns in Marker Space
So what we did is we identified these roi’s and then we trained a classifier based on the cyclic immunofluorescence data from each of those regions and then we used that to see if we could predict similar regions elsewhere in the specimen and in fact using that CyCIF data from these are distinct ROIs. We were able to retrieve areas very nicely that were, that looked exactly the same as the ones that were they were trained on, so normal predicted, normal glandular, predicted glandular, solid and mucinous. But we also notice as you can see here in this plot of shannon entropy, that there were a lot, that have high shannon entropy, that a lot of these regions are poorly mapped. So that was kind of interesting to us so we could definitely knew that disease relevant morphologies in H&E are encoded as hyper dimensional patterns in marker space. But we also noticed that some of this was more complex than we thought.
Most Regions of Tumor Compromise Mixtures of Morphological Classes with Continuous Molecular Gradients
And when we looked at these regions that were poorly predicted we saw that that’s because morphologically there were areas of transition. We had normal that was transitioning into more dysplastic glandular, we had mucinous areas that were transitioning into solid sheets of tumor, and we had glandular regions that were transitioning into solid. So we have this rather remarkable molecular plasticity that we see in H&E, but it’s also recapitulated in marker space so we see large transitions and gradients in E-cadherin and PCNA cytokeratins and interestingly, in oncogenic signaling in tumor suppressors, in epigenetic space and this is just a small snapshot of trimethylation of H3K27 but we see this across a whole range of epigenetic markers that these are amazingly rich gradient filled tumors that are, that do not have uniform phenotypic properties, almost reminiscent of gradients in developmental biology and now we can start to study what are driving these gradients. But it’s clear there’s a lot of heterogeneity.
Tumor Buds have an EMT Phenotype Molecular Gradients
Another point from this again this one sample but we see this across a whole range of colorectal cancer specimens, is that we can use our pathology driven review to find interesting biology as well. So there’s a property of a colorectal cancer at the invasive front where said cells are said to be budding, there are these tumor buds which you can see, which are hard to see on H&E actually but you can see them pretty nicely here stained by cytokeratin in white and they’re, by international consensus, a bud is a tumor is less than less than or equal to four tumor cells at the invasive front and this is associated with very poor prognosis in colorectal cancer. What we see from our three-dimensional reconstructions is that these are not really individual cells but actually they’re parts of finger-like tendrils that actually stem, that go all the way back into the tumor mass and you can see that here with this three-dimensional reconstruction. But interestingly, with the multiplex data, we can see that as before, these were known to be EMT, in an EMT like state and that’s what we see but these EMT states they don’t just start abruptly at the buds but they’re these long molecular transitions that go all the way back into the main tumor mass so they’re these continuous gradients, so for instance, E-cadherin if we look at tumor cells by cluster size proliferation are very low in the EMT state but they don’t just abruptly arise, they’re actually gradients that occur over long, long transitions and these are just a couple of the markers that we see but we see this with many markers and it’s pretty remarkable to start to see this now in both two dimensions and three dimensions and that takes us to this slide.
Multiplexed Data Adequate Tissue Sampling is Very Important
I’ll transition to Orion data in the next slide but I think this is an important consideration and as I showed you there these molecular gradients, these molecular transitions, if you look at the invasive front and the tumor environment and we map out interactions between keratin positive PD-L1 expressing tumor cells and exhausted T cells, these contour lines show an epicenter of those interactions. They’re not seen over here in this region that’s mucinous and they’re kind of moderately present but pretty weak in the more well-differentiated adenocarcinoma. So if you were to image or sample only this region, you’d get a very different understanding of the tumor than if you were to sample this region over here. So to explore this in more detail, we generated a tissue microarray from 16 whole tumor blocks. We put those cores into tissue microarray and then we what we did is we imaged using cyclic immunofluorescence the TMA. We also imaged the whole slides and then we computed in both the TMA and the whole slides the presence the frequency of these various individual marker states or double marker states or more.
And what we found is if we look at the whole slides, there is, you know for each marker there’s a fair amount of variability between slides, but it was its modest in some ways but, there is clearly variability between the frequencies of these populations in the 16 cases. If we look at the cores though from those 16 cases however, there’s much larger variability and then if we look at the cores from an additional 77 cases it kind of matches the variability we see with the cores and the 16 patients. So, over here is kind of our summary of this, so that measuring basic features of tissues, these markers here, may be subject to rather high sampling error included, that’s introduced when small fields of view are imaged or when small pieces of tissue are taken for imaging. So the sampling error in is greater in many cases than the true patient to patient variability and this is an area that I think our field needs to really explore and think hard about, but it’s an argument for why we think the whole slide imaging is going to be important. It’s also, I’m trying to make an argument here for why integration of our understanding with H&E and you know in 100 years of pathologists work on understanding the H&E is going to be very important for understanding the meaning of the molecular data that we’re getting.
Rapid Highly Multiplexed Immunoprofiling of Fixed Human Tissues by Orion Imaging
So in this regard, with this experience with cyclic immunofluorescence, we kind of got a sense of the pros and cons of the cyclic method and we stepped back and started thinking about what are the requirements for, what are the design features basically of optimal tissue imaging platforms and that would be compatible with pathology workflows both in the research setting, in the clinical translational setting, and hopefully at some point in the clinical setting. So these are the ten principles, basically you know we whole slide, highly multiplexed fluorescence imaging of tissues on glass slides is important so that we can get the entirety of the sample image and do an analysis on the entirety of the sample and this is a critical feature for digital pathology which in our survey of experts and the FDA’s guidance recommends using whole slide data. We’re looking for rapid, same-day, single pass, data acquisition rather than the cyclic sequential methods which can lead to some tissue degradation, which also just take a long time acquiring data from one slide using our method, one cycle is a day, we run 30 slides at a time or 40 using the HT3. But nonetheless, you know it does take about two weeks to collect a full data set, although it does take quite a while to understand it too so that two days, that two weeks is not, it’s not maybe prohibitive. We also you know, we believe in that it’s really important to do simultaneous channel acquisition in the range of 12 to 20 markers because this allows us to then enumerate immune cell types, tumor cells, and stroma and we can readily distinguish them and as I mentioned enumerate them and define their cell state.
On number four, the importance of imaging with subcellular resolution so that the data can be readily obtained so that the data can be readily obtained on single cells via segmentation and single cell analysis, and this permits also a study of critical sub-sailor events which is kind of a new frontier in some of the work that’s coming out of multiplex imaging. Number five is the removal of autofluorescence to assist in the detection of low expression proteins, for instance PD-L1, and we also feel that there’s a lot of information present in the autofluorescence that can also be very informative and useful particularly when assessing some stromal components. Number six, it’s obvious compatibility with formalin fixed and paraffin embedded FFP tissues. You know this is what we routinely use in pathology labs and it’s just one of the workhorses, but these methods should also be applicable and usable on frozen tissues. Number seven is bright field imaging of standard tissue stains such as H&E on the same tissue section. What’s really nice about this and I’ll show you a little bit in a while is that is then you can as I mentioned, integrate multiplex information and know-how and knowledge into the context of H&E but it also allows for combined learning approaches. And then another obvious point is availability of customizable and validated antibody panels both for human and for model organisms such as mouse, optical and mechanical stability, this is important for stitching together these large whole slide images from what can be hundreds of individual fields of view and then compatibility with H&E established image formats ome tiff, bio formats 6.0, and metadata standards. As I showed earlier, we have a metadata standard called mighty which is important for sharing data.
So, these are the design features and you know the Orion system really delivers on many of these. It offers 21 if not all of these, so it offers 21 channel, whole slide fluorescence imaging, staining rapidly whole slides in hours and not days. There’s a single staining procedure which is important for, just for technical reasons whether that’s in the research laboratory in a core, or in the clinical setting, it allows for high resolution imaging. We typically image using the 20x objective but you can also use other objectives as well but the 20x provides really great imaging data that allows for discrimination between subcellular compartments. Flexible panel design – RareCyte markets these as the StrataPlex reagents, FFPE compatibility as I mentioned, H&E imaging from the same slide using the same instrument after acquisition of the multiplex data again on the same section and then open file formats, RareCyte offers Artemis software for visualization and as I mentioned before our pipeline like MCMICRO can ingest this data and the data that the analysis that I’ll show you is all through the MCMICRO pipeline.
Whole Slide High-plex Imaging Data
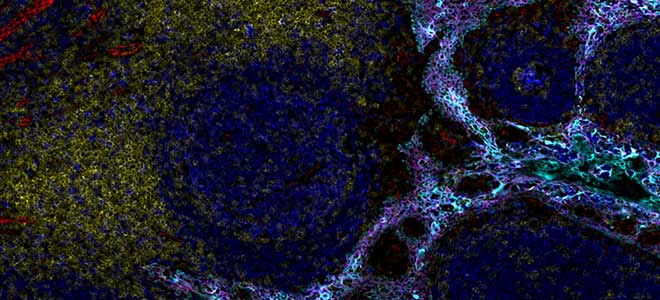
So for some pretty pictures this is a whole slide, high plex image of a tonsil, very characteristic of multiplex platforms right now. Uou can see really striking covering of the surface mucosa b cells marked by CD20 that are also obviously proliferative because of the terminal general centers and then there’s a mantle of in white of CD4 positivity and that’s marking the T helper cells. This is from a run using 17 plex Orion imaging and this was acquired in less than two hours and it’s rather large piece of tissue, those images.
Orion Tonsil Imaging
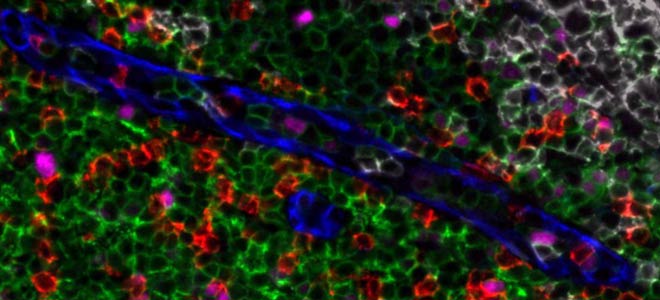
Zooming in to some of the data it’s you know, really pretty images here again the germinal center with proliferating B cells marked by CD20 and PCNA, mantle of CD4 T cells and a scattering of macrophages here in purple and blood vessels small capillary and venules that are marked by CD31 again, this is all from the same field of view. Instead of just PCNA we also have Ki67, PD-1 and the T follicular helper cells and CD3 co-marking the CD4 cells scattering here in the next panel over of CD8 cells, T cells as well and then a small number of FOXP3 positive cells in tonsil which is characteristic but in for instance in our imaging of colorectal cancer we see many CD4 positive FOXP3 positive cells and then just another basic view of co-expression of PD-L1 on macrophages and neighboring macrophages that don’t express PD-L1 but that expressed CD163.
Spatial Resolution of Overlapping Signals
So how does this happen, this is an example of the spectral resolution of overlapping signals, it’s one image using one of the lasers, excitation 555 emission 606, collecting data on five markers. So these are the markers unresolved sort of keratin macrophages T cell markers as well. These are images of Orion having extracted the overlapping signals into separate channels so you can see them individually here and then they’re merged in a pseudo color composite with the signal spectrally and spatially resolved.
Crosstalk Between Adjacent Channels
So, one key question has to do with crosstalk between channels, between adjacent channels basically quote unquote bleed through and obviously that happens we see that in the raw data as you can see here this is a correlation of pixel intensities between all the raw channel pairs and these are ordered by their emission wavelengths. There’s clearly bleed through from one you know, from one channel to the next but that’s removed after you know, by the Orion extraction. So for instance the PCNA channel does not have bleed through, it does not bleed through into the CD4 channel and the spatial correlations that you see the signal correlations that you see here are biological so for instance PCNA correlates with Ki-67, two proliferation markers we’ll see correlation between CD3 and CD4 and CD8, which are expected because they co-label, they co-segregate by labeling the same cells or similar cells. So, this is the type of extraction quality that you see and these are the images that correlate with this experiment. So for instance E-Cadherin here and PanCK are co-expressed, PD-L1 is expressed in a portion of the mucosal in tonsils so that you can see that co-localizing here and etc, etc. You can walk through these and be convinced that it is really very good and rapid extraction that’s happening with this method.
Image and Analyze Any Sample Type
I want to point out that you can image and analyze any sample type. These are just four examples of the types of samples that have been imaged to date. So as I mentioned shown in tonsils but skin is readily accessible, lung is as I mentioned there’s a minerva story available at images.rareyte.com that’s available also thymus etc, etc no tissue is inaccessible, both fixed and frozen.
This is an example of a colorectal cancer and it’s again whole slide image, rather large specimen. YTou can see here the normal colonic epithelium with scattered tertiary lymphoid structures and here in green marked by CD45 the muscularis mucosa is present here, the muscularis propria is present in red both marked by smooth muscle actin you can see the tumor as it forms abruptly here marked in pink slash purple, large tumor mass invading into the muscularis propria and through into the visceral fat and you can see these chorus yetis that of course from TMAs giving you a sense of how small of you get when you’re imaging TMAs relative to the entire tumor mass and just one of the benefits of whole slide imaging for exploring the various portions of this, whether it’s the part that’s invaded into the visceral fat, the part that’s invaded into muscularis or the molecular transitions that I was showing before in the main tumor mass.
Image and Analysis – Whole Slides
We for all the Orion data now we’ve run the data as I mentioned through MCMICRO and we can extract single cell data and this is a dimensionality reduction just using a simple t-SNE from the specimen I showed before just to give you just to show you that you know we can readily separate out the signal from the CD31 vascular cells from CD45 immune cells. PanCK overlaps really nicely with E-Cadherin here in it but it’s separate from CD45 and also separate from SMA CD68 positive macrophages co-label some well CD68 co-labels macrophages with CD163 but there are also distinct populations of each as well you can see here also FOXP3 co-localizes with a with a subset of the CD4 cells, CD4 and CD8 separate from one another. PD1 is present in a subset of the of the of the cytotoxic T cells and etc etc. You know, here’s another one that I’ll point out. It says the proliferation markers are predominantly in the tumor in this case but also a little bit present in some of the immune compartments too.
Image and Analysis – Tissue Microarrays
I also want to point out that analysis from the whole slide is analysis from tissue microarray. So this is a tissue microarray that the acquisition took roughly three hours using a 17-18 plex panel. This is from a quadruplicate cores of 93 different from 93 different patients, here’s just a just one image from one of the cores and again you know walking through we can readily extract the information at single cell level from the various markers as will be expected from this particular panel that was used.
Multimodality Data Fusion
And the other thing that I’d like to point out and I mentioned before is the ability to integrate H&E with multiplex data. So I showed you this sample earlier, this is the one that we did the three-dimensional reconstruction on but now this is imaged with Orion and when we compare the cell counts, the cell state counts from CyCIF and Orion it’s very similar which is reassuring to see, and one of the nice parts of Orion is that we can move between multiplex space and H&E space because it’s all on the same section. And again, I don’t hope this hopefully this is running now so you can see this is a tool that our colleagues, Hanspeter Pfister and Robert Kruger at the Harvard school of engineering and applied sciences generated, which allows us to flip between multiple modalities, in this case H&E and CyCIF but you can flip through RNA data as well and I’m sorry in Orion in this case. So what’s neat here again is that you not only get the actual H&E on the same section that was image but you can then use for instance, molecular labels to label features, label cells in the H&E.
Image and Analysis of Lung Primary and Brain Metastases
What I’d like to show in this last slide is one another experiment that we did where we took 10 different matched pairs of lung cancer that the matches are between the primary tumor and the brain metastasis and we use Orion to profile the primary and the met from each of these into these patients and then quantify the number of cells. This is very basic analysis and I’m not showing you any of the spatial analysis that we’ve done but just in terms of enumeration of cell types and what we find when we compare the primaries with the mets is that there’s a there’s a rewiring of the immune microenvironment between the two sites. Lung tumors in the lung have much higher B cell populations, fibroblasts perhaps not surprising as well as well as T cells, FOXP3 cells, and CD8s than we see in the brain, and in the brain when we look at the tumor compartment we see a burst of proliferation so these are more proliferative in the brain than they are in the lung. But this is just an example of how we’re starting to use this type of information.
Multiplexed Tissue Imaging to Reveal the Spatial Biology of Cancer
So in conclusion it’s a great era now I think in pathology. We have these remarkable new tools for quantitative spatial analyses. I showed you some data from CyCIF and from Orion we have an expanded capacity now for biomarker discovery with quantification and spatial analyses in human clinical tissues and also mouse models. Orion, it provides a multiplex method that’s compatible with pathology workflows so you can use a standard glass microscope slide, FFPE tissues. This is a single staining process with standard immunofluorescence protocols very familiar to our lab and many labs out there. One whole slide scan gives you up to 20 markers. You can do analysis with Artemis which is the RareCyte software but also the data that comes off the Orion platform is compatible with third-party and open-source software like MCMICRO which I showed. And you know in tissue analyses they’re absolutely going to be essential for patient management and optimal development of single and combination therapies and exploring cancer but also infectious disease. We’ve imaged tuberculosis we’ve imaged covid and other infectious processes which really feel strongly that in our generation, I’m a neuropathologist the brain is it’s a pretty big organ and to get a sense of the pathology in the brain, imaging one section from the hippocampus is probably not enough, when we review cases for let’s say a dementia workup, we frequently take 20 whole slides so that’s something that I can imagine is going to be also important in characterizing the pathology that’s going on in neurodegeneration as well.I t’s imaging large slides and large numbers of slides from individual patients that are part of larger cohorts so a platform that scales is going to be very important I think for some of that discovery work.
I want to thank our colleagues at the RareCyte team. We have a really wonderful collaboration with them on our STTR and SBIR and obviously our team members at the LSP and our funding sources the NIH, NCI for supporting us through H10 and CSBC and also the SBIR and STTR as well as Ludwig cancer research.
So what I’d like to do now is just put up this thank you slide maybe what I’d point out lastly is this is this is actually some of the autofluorescence internal and external elastic lamina in a blood vessel and artery here. So you can get a sense of how we can even start to use some of the autofluorescence but I’ll stop talking about the data and the slides and to say thank you and take any questions now and also point out that we’re glad to answer questions either directly to RareCyte or also I’m also happy to answer any questions you might have.
Q&A for Cell and Tissue Imaging
Thank you very much Dr. Santagata. We do have several questions, our first question is from AI:
Does H10 have the database of protein expressions in multiple organ tissues?
That’s a very good question, so our goal is to is to characterize data across a whole range of different cancers. Hub map is focused on normal tissues and we work closely with some of the investigators there to, in that work, we’re focused more on tumors so we have a data portal both hub map has a data portal and we have one in H10 and all of that data as it gets collected is being uploaded there and we’re building out the tools to share some of this multiplex data in the H10 the viewers like Minerva and other viewers as well so that you should be able to scan through images and ultimately there will be a fusion of the capacity for instance of CBIO portal type capacity with these image viewers so that you’ll be able to view multiplex data alongside the genomic data for specific specimens and that gets to the point where we need to scale because to do this at the scale that’s needed to make the insights that I think we need lots of samples.
All right perfect. And we have a comment and a question from Simon, beautiful pictures. It is increasingly possible to generate image images such as these but making biological sense of the cell interactions is a bottleneck made harder by increasing numbers of markers and cell types.
Have you made any progress with understanding the biological significance of the cellular interactions you’re able to identify?
Yeah, that’s a great question and there’s no doubt that the real bottleneck, one of the real bottlenecks now that I think Orion has solved some of our issues with acquiring data even faster is computational and not just computational but it’s understanding so I started with that slide about the LSP because I really feel that that multi-disciplinary environment is really important for extracting biologically, biological information. I showed that we’re trying to do this in the context of H&E because we want to we don’t want to just understand the data in high dimensional space, but we want to understand it in the context of morphology. I showed the example of CD73 and 39 where we’re trying to understand these patterns and this is one that makes sense but it’s nice to see that these ecto enzymes are partnering not just in their enzymatic activity but actually in space to do this but I think this is the beginning obviously of a whole new era and the type of questions you’re asking are you know are very pertinent that the MCMICRO pipeline was developed to make that easier. Minerva is developed to showcase the data and to share it and mighty was developed to share data in repositories so that it can be mined by other groups and not just by the group that that generates it kind of like GEO is for genomic data so great question but I think there’s a lot of progress and hopefully groups like yours and hopefully ours will also and many others on this call will start to make progress and lead the way forward.
Perfect thank you, our next question is from Ratna Bali.
Is it possible to use this multiplex tissue imaging technique for samples much thicker than a sample on a glass slide, say a more 3D sample and if so how does the resolution compare?
That’s great so, this is one that I think I will have to defer to RareCyte because I assume the answer is yes but we haven’t done optical sectioning using Orion extraction which I think must be technically possible. What we are doing using other approaches is to do optical sectioning of let’s say 5 and 10 micron sections so you can develop a three-dimensional view at high resolution of the data within that five micron section instead of pouring it all together into you know into one projection. The reason why that’s important it’s many fold right there’s a lot of really interesting interactions that are happening that are not always easily pulled out of standard resolution imaging so this polarization of molecules. We right now we treat cells as is pretty poor flow cytometry in a way right where we’re integrating all of the information into and assign it to one cell but there’s a lot of morphological information there as well as distribution of molecules. So that’s on us as computationalists to start to extract more but maybe I’ll have RareCyte get back to you with maybe some of their thinking about how to do this in, with greater z then than I’m talking about.
All right our next question is from JV.
Can you comment on the future opportunities to leverage next-gen computational biology methods including AI on multiplexed tissue imaging impacting patient outcomes in cancer?
Yeah so I think that we’re going to really need to integrate multiplex data with some of the innovations that are happening on H&E analysis that’s one way where we I think we can start to really use next generation computation and also with radiology we’re starting to map these we historically over the last decade we’ve been mapping samples in the context of the of the MRI where the you know the space the three-dimensional space in the patient where the sample was taken so I think we’ll have to do further and further data integration with multiplexing in H&E which as I mentioned the Orion system really nicely does but also with other modalities and with space within the patient. I also think that we’re right now we’re on doing single cell segmentation and as a pathologist you know when I’m scanning through slides I’m not looking at single cells, I’m getting like this global picture we’re still trying to learn what we see as pathologists when we make our diagnostic calls and I think AI approaches, next gen AI, will probably not necessarily work at a single cell level but will take expression more globally, regionally and start to understand samples in that way so I think there’s a great opportunity there.
All right thank you, I see we have time for just one more question, we have a question from MC:
Cool stuff but it seems quite similar to chip cytometry. How does this differ?
I’m not no I understand cytometry so I’m probably not the right person to tell you how it differs you know one aspect of this is that they’re you know just now any one center can have multiple technologies to do comparisons so these types of head-to-head comparisons are part of the H10 effort. We have we for instance that’s colorectal cancer specimen that I showed you from the cooperative human tissue network, we’ve distributed that to eight different centers that each are doing a different technology and we’re accumulating, we’re collecting that data and performing joint analyses to do sort of head-to-head comparisons to see where one method might be better and others might not, might be worse or whatever you know however it is when you want to leverage a particular method because each of these has their pros and their liabilities.
Thank you so much we do have several more questions but we have run out of time so we’ll be forwarding those questions along to our presenter and RareCyte to get the answers for you and you’ll receive those answers via email. I would like to thank our presenter for today Dr. Sandro Santagata I’d like to thank RareCyte for sponsoring today’s event I also want to take this opportunity to thank those of you who came and spent this time with us we’re very grateful you chose to be with us and we hope you found the information helpful and exciting on behalf of Cambridge innovation institute’s global web symposia series thank you all so very much for attending and have a great rest of your day, bye.